One of the essential factors the COVID-19 virus needs to enter a host is a receptor on a human cell — a place where the universally recognized spike protein can latch onto the cell surface, pierce it, disgorge its infectious contents, and replicate.
Without a receptor, there is no replication. Without replication, there is no infection.
Researchers in Princeton University’s Department of Chemistry and the Department of Molecular Biology have used a cellular mapping technology called µMap, introduced just two years ago by the MacMillan Lab, to uncover eight previously unknown entry points of interest for the spike protein.
Four of them, researchers found, are functionally important for viral entry.
The research was published earlier this month in the Journal of the American Chemical Society (JACS). It could expand the suite of tools used to fight the virus, particularly as it mutates and evolves ways to evade vaccines.
Saori Suzuki, a postdoc in the Ploss Lab and lead author on the JACS paper; and Alexander Ploss, professor of Molecular Biology.
The collaborative project was begun at the height of pandemic uncertainty two years ago under Alexander Ploss, a leading virologist and professor of molecular biology, and David MacMillan, the James S. McDonnell Distinguished University Professor and a Nobel laureate in chemistry.
Scientists have known since the SARS-CoV-1 virus appeared in 2003 that its primary viral entry receptor was an enzyme called angiotensin-converting enzyme 2, or ACE2. This enzyme was confirmed in 2020 as the same receptor for SARS-CoV-2, the virus that causes COVID-19.
But the Princeton project started with the assumption that ACE2 was not the only story.
“We did know that there are certain host molecules that this virus absolutely depends on to enter into lung cells to cause the infection, and one of these molecules is called ACE2,” said Ploss. “So we basically said, okay, let’s see if there’s more out there. We looked for immediate binders.
“But as you can imagine, the entry process is complex. The virus attaches to something and then it still has to pass through the cell membrane to get into a cell, and along this way it may interact with other host factors. I don’t want to say everything is dictated by viral entry. Obviously, there are a number of equally essential processes within the cell after the virus has entered that can influence disease severity.

Steve Knutson, a postdoctoral research fellow in chemistry
“But it’s obviously the first key step. If the virus can’t get in, it’s game over.”
Steve Knutson, a co-author on the paper and a postdoctoral research fellow in the MacMillan Lab, added: “While the discovery of ACE2 as the major receptor was a huge milestone, it certainly doesn’t tell the whole story of COVID pathology. Biology can be inherently promiscuous, and we guessed correctly that the SARS-CoV-2 spike protein interacts with multiple host cell proteins for entry.”
He added that investigations like this one are a “perfect” research fit for the µMap technology.
The spike as µMap antenna
µMap, or micromap, is a proximity labeling technology that identifies protein and enzyme “neighbors” on a cell’s surface. It uses a photocatalyst — a molecule that, when activated by light, spurs a chemical reaction — to flag these spatial relationships by generating a marker that tags molecular neighbors.
In this work, researchers used the spike protein itself as the marker or “antenna” to tag all the receptor sites in the vicinity of ACE2.
“Alex had this great instinct that there’s other things apart from ACE2 that could allow you to think about infectivity,” said MacMillan. “So what we did was put this photocatalyst — and we call them antennas — on the spike protein, so that whenever it binds to things on the cell near ACE2, this little antenna absorbs the photonic energy, the light.
“But it can’t give that energy away over long distances. It can only give it to what’s close by. The molecule that’s free floating has to basically encounter it within two nanometers,” MacMillan added. “So we know what’s next to it. We know what’s interacting with it.”
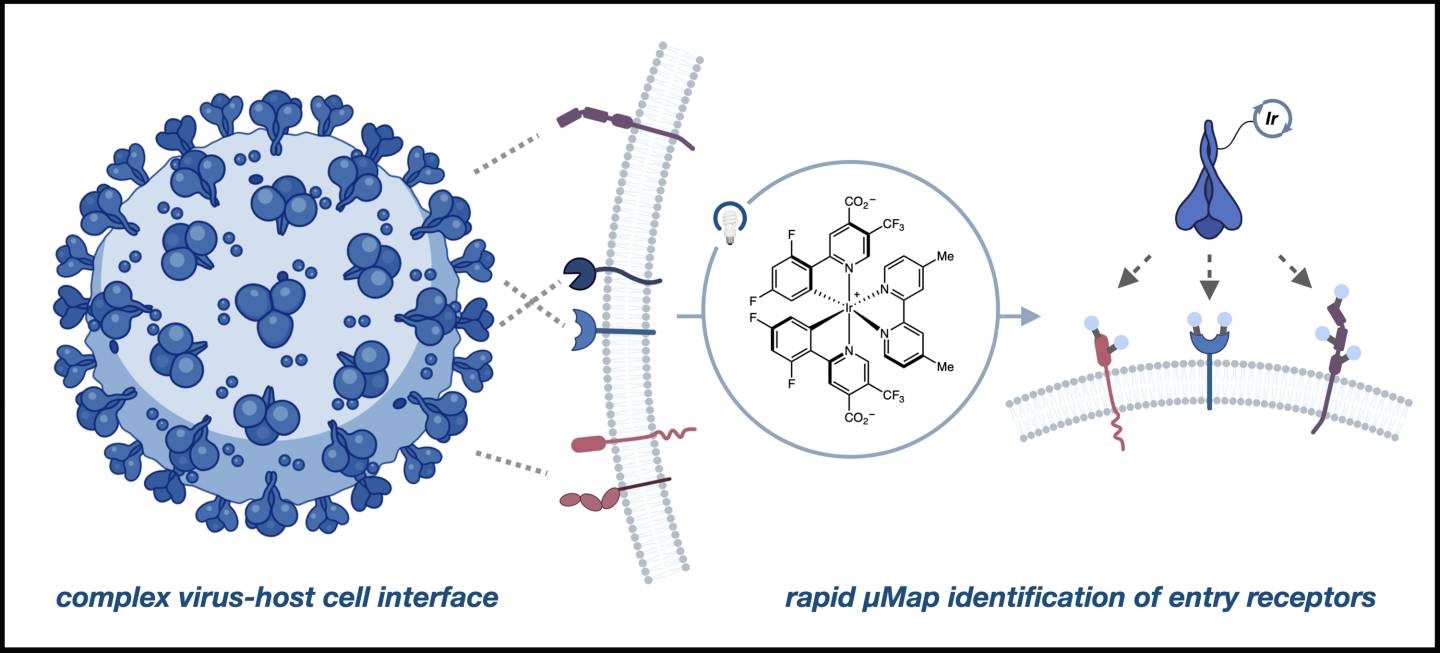
The host cell and virus interface can be complex and involve many proteins, but µMap enables direct interrogation of functionally important interactions.
After the technology identified eight novel receptors that interacted with the spike protein, scientists characterized them using a virus pseudoparticle. (A pseudoparticle mimics viral entry but does not carry the genetic material to spread the virus.) They then isolated four entry factors worthy of further investigation.
“The pseudoparticle system allows us to uncouple viral uptake and study the entry process from everything downstream of the infectious cycle,” said Ploss. “If you’re looking for the impact of certain host factors on entry, you want to see that you can study it independently of replication. So here, we’re basically introducing a reporter gene into the cell and can then quantify how efficiently entry has taken place.”
MacMillan said that further work was needed to ultimately determine the function of receptors, but researchers wonder if they might carry a clue to disease severity.
“We cannot say all eight factors are related to SARS-CoV-2 entry,” said Saori Suzuki, an associate research scholar in the Ploss Lab. “Four factors out of eight were outstanding by virological assessment. We need to assess more and evaluate more precisely.
“In the next step, we need to assess how these factors support ACE2 for virus entry and whether newly emerging viral variants utilize the same set of factors.”
“Photochemical Identification of Auxiliary Severe Acute Respiratory Syndrome Coronavirus 2 Host Entry Factors Using µMap,” by by Saori Suzuki, Jacob B. Geri, Steve D. Knutson, Harris Bell-Temin, Tomokazu Tamura, David F. Fernández, Gabrielle H. Lovett, Nicholas A. Till, Brigitte L. Heller, Jinchao Guo, David W. C. MacMillan and Alexander Ploss, appeared in the Sept. 1 issue of the Journal of the American Chemical Society (DOI: 10.1021/jacs.2c06806.) The research was funded in part by grants from the National Institutes of Health (R01AI138797, R01AI107301, R01AI146917, and R01AI153236 to A.P. and a component of UL1TR003017), a Burroughs Wellcome Fund Award for Investigators in Pathogenesis (101539 to A.P.), Princeton COVID-19 research funds through the Office of the Dean for Research, and the NIH National Institute of General Medical Sciences (R35-GM134897-02).