For most of our evolutionary history — for most of the time anatomically modern humans have been on Earth — we’ve shared the planet with other species of humans. It’s only been in the last 30,000 years, the mere blink of an evolutionary eye, that modern humans have occupied the planet as the sole representative of the hominin lineage.
But we carry evidence of these other species with us. Lurking within our genome are traces of genetic material from a variety of ancient humans that no longer exist. These traces reveal a long history of intermingling, as our direct ancestors encountered — and mated with — archaic humans. As we use increasingly complex technologies to study these genetic connections, we are learning not only about these extinct humans but also about the larger picture of how we evolved as a species.
Joshua Akey(Link is external), a professor in the Lewis-Sigler Institute for Integrative Genomics(Link is external), is spearheading efforts to understand this larger picture. He calls his research method genetic archaeology, and it’s transforming how we’re learning about our past. “We can excavate different types of humans not from dirt and fossils but directly from DNA,” he said.
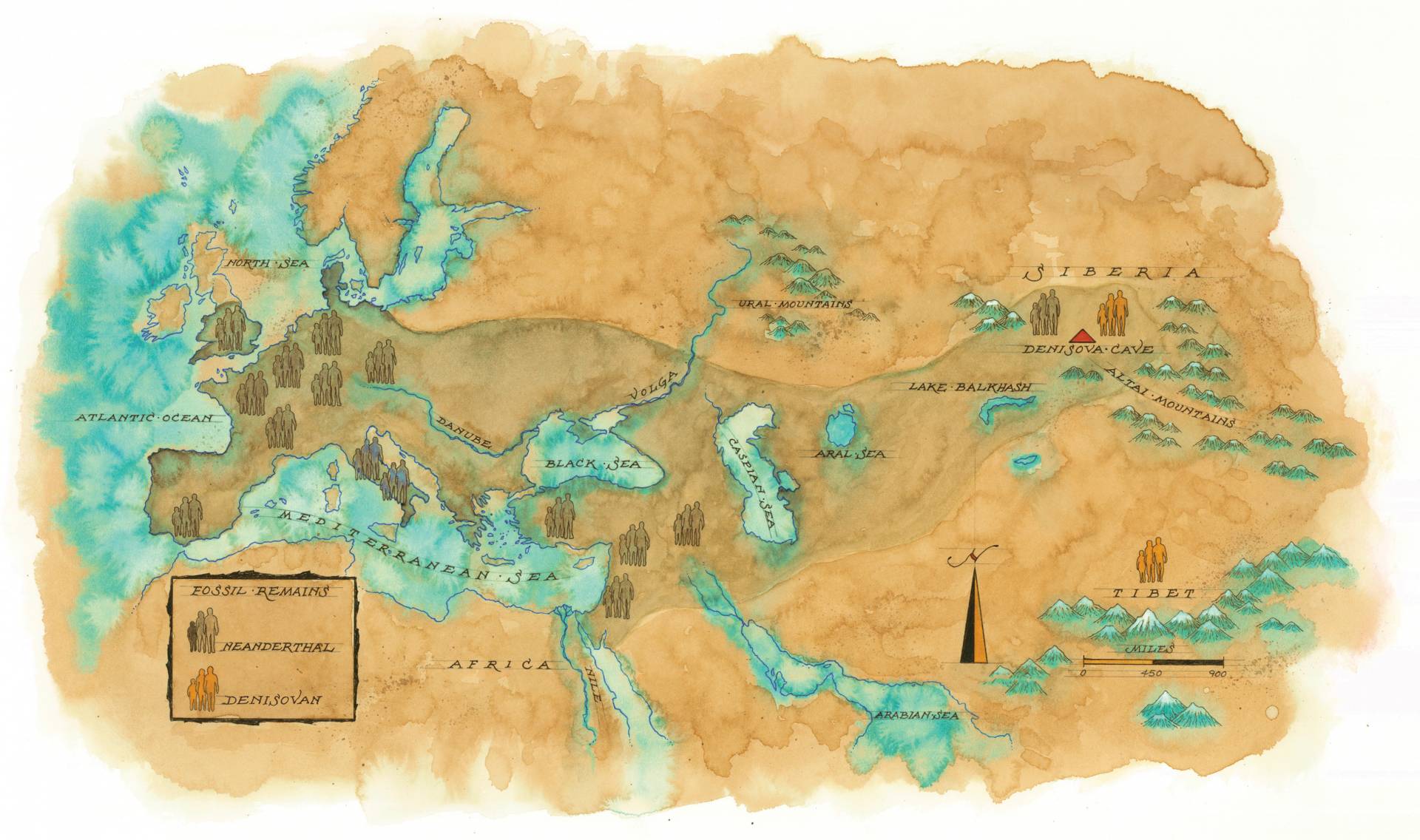
Joshua Akey, a professor in the Lewis-Sigler Institute for Integrative Genomics, uses a research method he calls genetic archaeology to transform how we’re learning about our past. Fossil evidence illustrates the spread of two long-extinct hominin species, Neanderthals and Denisovans. Modern humans carry genes from these species, indicating that our direct ancestors encountered and mated with archaic humans.
Combining his expertise in biology and Darwinian evolution with computational and statistical methods, Akey studies the genetic connections between modern humans and two species of extinct hominins: Neanderthals, the classical “cave men” of paleoanthropology; and Denisovans, a recently discovered archaic human. Akey’s research divulges a complex history of the intermixing of early humans, indicative of several millennia of population movements across the globe.
“There’s often a divide between the researchers who go out and collect exotic samples and the researchers who do really creative theory and data analysis, and he’s done both,” said Kelley Harris, a former colleague of Akey’s who is now an assistant professor of genome sciences at the University of Washington.
Like many of us, Akey has long been interested in how the human species evolved. “People want to learn about their past,” he said. “But even more than that, we want to know what it means to be human.”
This curiosity followed Akey throughout his schooling. During his graduate work at the University of Texas Health Science Center at Houston in the late 1990s, he looked at how contemporary humans in different parts of the world were genetically related to one another, and used early gene sequencing methods to try to understand these relationships.
Gene sequencers are devices that determine the order of the four chemical bases (A, T, C and G) that make up the DNA molecule. By determining the order of these bases, analysts can identify the genetic information encoded in a strand of DNA.
Since the 1990s, however, gene sequencing technology has progressed dramatically. A new technology known as next-generation sequencing came into use around 2010 and allowed researchers to study a very large number of genetic sequences in the human genome. It took 10 years to sequence the first human genome, but these new machines get whole genome sequence data from thousands of individuals in only a matter of hours. “When next-generation sequencing technology started to become the dominant force in genetics,” Akey said, “that completely changed the entire field. It’s hard to overstate how dramatic this technology has been.”
The scale of the data that now can be analyzed has allowed researchers to address a whole slew of new questions that would not have been possible with the previous technology.

Joshua Akey and his team use gene-sequencing technologies to reveal new information about archaic human lineages as well as our own evolutionary history.
One of these questions is the relationship between modern humans and archaic humans, such as Neanderthals. In fact, this question fostered a vigorous debate about whether modern humans carried genes from Neanderthals. For many years, the opinions of researchers — both pro and con — ticked back and forth like a metronome.
Gradually, however, a few researchers — including geneticists Svante Pääbo of the Max Planck Institute in Germany and his colleague Richard (Ed) Green of the University of California-Santa Cruz — began to demonstrate strong evidence that, indeed, there had been gene flow from Neanderthals to modern humans. In a 2010 paper, these researchers estimated that people of non-African ancestry had about 2% Neanderthal ancestry.
Neanderthals lived in a wide geographical swath across Europe, the Near East and Central Asia before dying out around 30,000 years ago. They lived alongside anatomically modern humans, who evolved in Africa some 200,000 years ago. The archaeological record shows that Neanderthals were adept at making stone tools and developed a number of physical traits that uniquely adapted them to cold, dark climates, such as broad noses, thick body hair and large eyes.
Following on the heels of Pääbo and Green’s Neanderthal research, Akey and a colleague, Benjamin Vernot, published a paper in Science looking at recovering Neanderthal sequences from the genome of modern humans. Geneticist David Reich of Harvard University published a similar paper in Nature, and, together, the two papers provided the first data employing the modern genome to investigate our link with Neanderthals.
Using the genetic variation in contemporary populations to learn about things that happened in the past involves scrutinizing the modern human genome for gene sequences that display traits expected to have been inherited from a different type of human. Akey and his colleagues then take those sequences and compare them to the Neanderthal genome, looking for a match.
Using this technique, Akey has been able to uncover a rich human legacy of genetic interconnections on a scale previously unconceived. As stated, while the available evidence suggests that non-Africans carry about 2% of Neanderthal genes, Africans, who were once believed not to have any connections with Neanderthals, actually have approximately 0.5% Neanderthal genes. Researchers have further discovered that the Neanderthal genome has contributed to several diseases seen in modern human populations, such as diabetes, arthritis and celiac disease. By the same token, some genes inherited from Neanderthals have proven beneficial or neutral, such as genes for hair and skin color, sleep patterns and even mood.
Akey has also discovered genetic fingerprints that suggest our human ancestry contains species about which we know nothing or very little. The Denisovans are a case in point. An archaic form of human, they coexisted with anatomically modern humans and Neanderthals and interbred with both before going extinct. The first evidence of their existence came in 2008 when a finger bone was discovered in Denisova Cave in the remote Altai Mountains of southern Siberia. At first the bone was assumed to be Neanderthal because the cave contained evidence of these species. Consequently, it sat in a museum drawer in Leipzig, Germany, for many years before it was analyzed. But when it was, the researchers were dumbfounded. It wasn’t a Neanderthal — it was a hitherto unknown type of ancient human. “The Denisovans are the first species ever identified directly from their DNA and not from fossil data,” Akey said.
Since that time, continued genetic work — much of it conducted by Akey and his colleagues — has established that the closest living relatives of Denisovans are modern Melanesians, the inhabitants of the Melanesian islands of the western Pacific — places such as New Guinea, Vanuatu, the Solomon Islands and Fiji. These populations carry between 4% and 6% of Denisovan genes, though they also carry Neanderthal genes.
Examples like this highlight one of the main features of our human lineage, Akey said, that admixture has been a defining feature of our history. “Throughout human history there’s always been admixture,” Akey said. “Populations split and they come back together.”

While there remains a lot of debate about the Denisovans, Akey believes they most likely were closely related to Neanderthals, perhaps an eastern version who split off from the latter sometime around 300,000 or 400,000 years ago. Recently, genetic analysis of fossils from Denisova Cave has uncovered evidence of an offspring between a Neanderthal woman and a Denisovan male. The offspring was a female who lived approximately 90,000 years ago. By looking at this genetic trail, Akey and other researchers have been able to piece together a fascinating story of human evolution — one that is promising to rewrite our understanding of early human origins.
But there’s so much more to discover, Akey said. “Even though we have sequenced probably 100,000 genomes already, and we have pretty sophisticated tools for looking at that variation, the more we think about how to interpret genetic variation, the more we find these hidden stories in our DNA,” he said.
This article was originally published in the University’s annual research magazine Discovery: Research at Princeton(Link is external).