A modern pygmy population evolved short stature independently of the extinct pygmy species that lived in the same region thousands of years earlier, report Princeton’s Serena Tucci, Joshua Akey and an international team of researchers. In this illustration, the modern village is shown at left; in the center, a modern pygmy wearing the traditional head covering and clothing is juxtaposed against the face of a Homo floresiensis reconstruction; at right, pygmy elephants play in the cave where the fossils were found.
Two pygmy populations on the same tropical island. One went extinct tens of thousands of years ago; the other still lives there. Are they related?
It’s a simple question that took years to answer.
As no one has been able to recover DNA from the fossils of Homo floresiensis (nicknamed the “hobbit”), researchers had to create a tool for finding archaic genetic sequences in modern DNA.
The technique was developed by scientists in the lab of Joshua Akey(Link is external), a professor of ecology and evolutionary biology(Link is external) and the Lewis-Sigler Institute for Integrative Genomics(Link is external) at Princeton University.
“In your genome — and in mine — there are genes that we inherited from Neanderthals,” said Serena Tucci(Link is external), a postdoctoral research associate in Akey’s lab. “Some modern humans inherited genes from Denisovans [another extinct species of humans], which we can check for because we have genetic information from Denisovans.
“But if you want to look for another species, like Floresiensis, we have nothing to compare, so we had to develop another method: We ‘paint’ chunks of the genome based on the source. We scan the genome and look for chunks that come from different species — Neanderthal, Denisovans, or something unknown.”
She used this technique with the genomes of 32 modern pygmies living in a village near the Liang Bua cave on Flores Island in Indonesia, where H. floresiensis fossils were discovered in 2004.
“They definitely have a lot of Neanderthal,” said Tucci, who was the first author on a paper published in the Aug. 3 issue(Link is external) of the journal Science that detailed their findings. “They have a little bit of Denisovan. We expected that, because we knew there was some migration that went from Oceania to Flores, so there was some shared ancestry of these populations.”
But there were no chromosomal “chunks” of unknown origins.
“If there was any chance to know the hobbit genetically from the genomes of extant humans, this would have been it,” said Richard “Ed” Green, an associate professor of biomolecular engineering at the University of California-Santa Cruz (UCSC) and a corresponding author on the paper. “But we don’t see it. There is no indication of gene flow from the hobbit into people living today.”
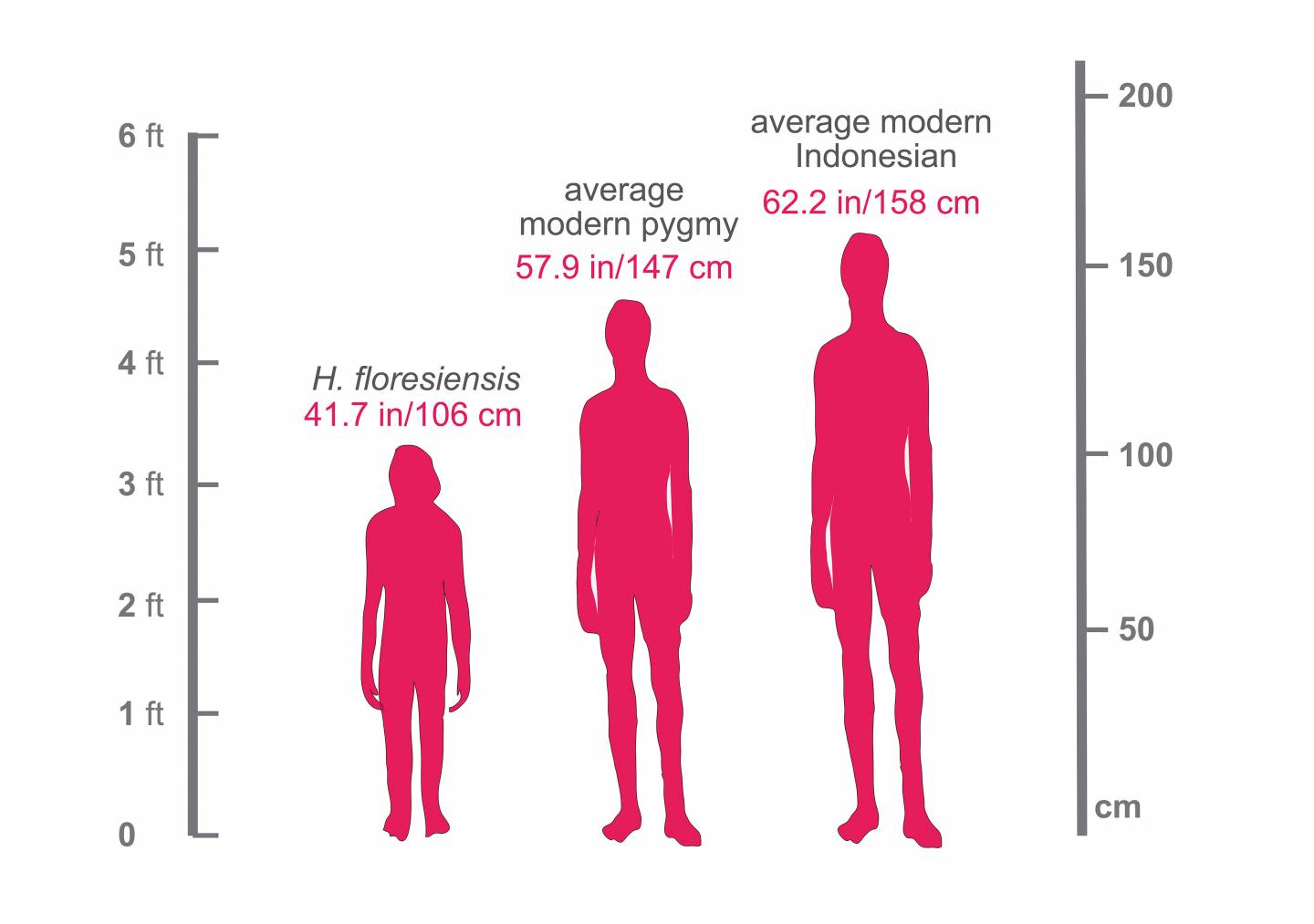
This figure shows the relative heights of a modern Indonesian (5’ 2), a modern pygmy living on the island of Flores (4’ 10) and Homo floresiensis (3’ 5, which is the height of an average American 4-year-old).
The researchers did find evolutionary changes associated with diet and short stature. Height is very heritable, and geneticists have identified many genes with variants linked to taller or shorter stature. Tucci and her colleagues analyzed the Flores pygmy genomes with respect to height-associated genes identified in Europeans, and they found a high frequency of genetic variants associated with short stature.
“It sounds like a boring result, but it’s actually quite meaningful,” Green said. “It means that these gene variants were present in a common ancestor of Europeans and the Flores pygmies. They became short by selection acting on this standing variation already present in the population, so there’s little need for genes from an archaic hominin to explain their small stature.”
The Flores pygmy genome also showed evidence of selection in genes for enzymes involved in fatty acid metabolism, called FADS enzymes (fatty acid desaturase). These genes have been associated with dietary adaptations in other fish-eating populations, including the Inuit in Greenland.
Fossil evidence indicates H. floresiensis was significantly smaller than the modern Flores pygmies, standing about 3.5 feet tall (106 centimeters), while modern pygmies average about 15 inches taller (145 centimeters). Floresiensis also differed from H. sapiens and H. erectus in their wrists and feet, probably due to the need to climb trees to evade Komodo dragons, said Tucci.
Dramatic size changes in animals isolated on islands is a common phenomenon, often attributed to limited food resources and freedom from predators. In general, large species tend to get smaller and small species tend to get larger on islands. At the time of H. floresiensis, Flores was home to dwarf elephants, giant Komodo dragons, giant birds and giant rats, all of which left bones in the Liang Bua cave.
“Islands are very special places for evolution,” Tucci said. “This process, insular dwarfism, resulted in smaller mammals, like hippopotamus and elephants, and smaller humans.”
Their results show that insular dwarfism arose independently at least twice on Flores Island, she said, first in H. floresiensis and again in the modern pygmies.
“This is really intriguing, because it means that evolutionarily, we are not that special,” she said. “Humans are like other mammals; we are subject to the same processes.”
This research involved a large international collaboration. Green of UCSC and Akey of Princeton were the senior principal investigators, and Tucci is first author on the paper. Herawati Sudoyo and Gludhug Purnomo of the Eijkman Institute for Molecular Biology in Jakarta, Indonesia, oversaw the collection of samples for genome sequencing from the Flores pygmies, and Purnomo assisted with sample processing at UCSC.
Other co-authors include Peter Visscher at the University of Queensland, Australia; Samuel Vohr at UCSC; Rajiv McCoy(Link is external) at Princeton; Benjamin Vernot at the Max Planck Institute for Evolutionary Anthropology in Leipzig, Germany; Matthew Robinson at the University of Lausanne, Switzerland; Chiara Barbieri at the Max Planck Institute for the Science of Human History in Jena, Germany; Brad Nelson, Wenqing Fu and Evan Eichler at the University of Washington, Seattle; and Guido Barbujani at the University of Ferrara, Italy.
“Evolutionary history and adaptation of a human pygmy population of Flores Island, Indonesia(Link is external),” by Serena Tucci, Samuel Vohr, Rajiv McCoy, Benjamin Vernot, Matthew Robinson, Chiara Barbieri, Brad Nelson, Wenqing Fu, Gludhug Purnomo, Herawati Sudoyo, Evan Eichler, Guido Barbujani, Peter Visscher, Joshua Akey and Richard Green (DOI:10.1126/science.aar8486), published in the Aug. 3 issue of Science, was supported by the National Institutes of Health (R01-GM110068), the Searle Scholars Program, the Gordon and Betty Moore Foundation, the Lewis and Clark Fellowship for Exploration and Field Research (American Philosophical Society), the University of Ferrara-Italy Young Researcher Fellowship program, the European Research Council (ERC-2011-AdG_295733), the Australian Research Council (DP160102400), the Australian National Health and Medical Research Council (1078037 and 1113400), and the Ministry of Research and Technology of the Republic of Indonesia.