You might have seen Hollywood stars in “motion capture” suits, acting in full-body costumes peppered with sensors that let a computer transform them into a Hulk(Link is external) or a dragon(Link is external) or an enchanted beast(Link is external).
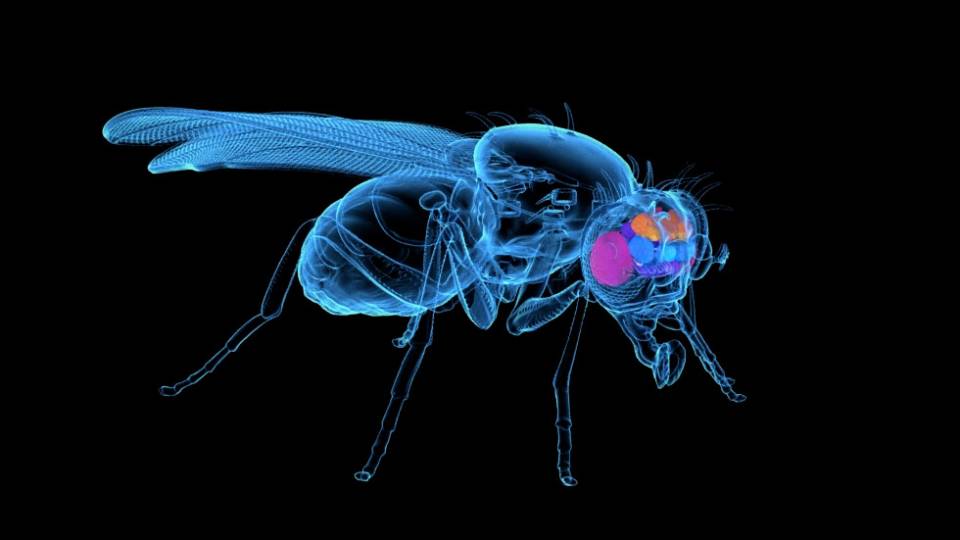
Now, a collaboration between the labs of Princeton professors Mala Murthy(Link is external) and Joshua Shaevitz(Link is external) has gone a step further, using the latest advances in artificial intelligence (AI) to automatically track animals’ individual body parts in existing video.
Their new tool, LEAP Estimates Animal Pose (LEAP), can be trained in a matter of minutes to automatically track an animal’s individual body parts over millions of frames of video with high accuracy, without having to add any physical markers or labels.
“The method can be used broadly, across animal model systems, and it will be useful to measuring the behavior of animals with genetic mutations or following drug treatments,” said Murthy, an associate professor of molecular biology(Link is external) and the Princeton Neuroscience Institute(Link is external) (PNI).
The paper detailing the new technology(Link is external) will be published in the January 2019 issue of the journal Nature Methods, but its open-access version(Link is external), released in May, has already led to the software being adopted by a number of other labs.
When the researchers combine LEAP with other quantitative tools developed in their labs, they can study what they call “the language of behavior” by observing patterns in animal body movements, said Shaevitz, a professor of physics(Link is external) and the Lewis-Sigler Institute for Integrative Genomics(Link is external).
“This is a flexible tool that can in principle be used on any video data,” said Talmo Pereira(Link is external), a PNI graduate student who is the first author on the paper. “The way it works is to label a few points in a few videos and then the neural network does the rest. We provide an easy-to-use interface for anyone to apply LEAP to their own videos, without having any prior programming knowledge.”
When asked if LEAP worked as well on large mammals as it did on the flies(Link is external) and mice(Link is external) that made up most of the initial subjects, Pereira promptly created a motion-tagged video of a giraffe taken from the live feed(Link is external) from Mpala Research Centre(Link is external) in Kenya, a field research station for which Princeton is managing partner.
“We took a video of a walking giraffe from the Mpala research station … and labeled points in 30 video frames, which took less than an hour,” Pereira said. “LEAP was then able to track motion from the entire rest of the video (roughly 500 frames) in seconds.”
Previous efforts to develop AI tools that could track human motion have relied on large training sets of manually annotated data. That allowed the software to work robustly on diverse kinds of data, with vastly different backgrounds or lighting conditions.
“In our case, we optimized similar methods to work on data collected in a laboratory setting, in which conditions are consistent across recordings,” said Murthy. “We built a system that allows the user to choose a neural network appropriate for the kind of data that the user collected rather than being constrained by what other researchers or companies have worked on.”

The software began as the senior thesis project of Diego Aldarondo of the Class of 2018, seen here at Class Day with his adviser, Mala Murthy.
This project arose from a unique collaboration between a senior thesis student in the Murthy lab(Link is external), Diego Aldarondo of the Class of 2018, and his graduate student mentor, Pereira, who is jointly advised by Murthy and Shaevitz.
“Diego was exploring the use of deep neural networks for annotating animal behavioral data via one of his computer science classes at Princeton, and over late-night chats in the lab with Talmo, he realized that these methods could be powerfully applied to their own data: videos of fruit flies interacting during their courtship ritual,” said Murthy. “The collaboration took off from there, and it was incredible fun to work together — Diego and Talmo showed how effective these AI methods can be.”
The work has great potential outside of neuroscience as well, said Monica Daley, a senior lecturer at the Structure and Motion Laboratory of the Royal Veterinary College in the United Kingdom, who was not involved in this research.
“Much of my research aims to understand how animals move effectively under different terrain and environmental conditions,” Daley said. “One of the biggest ongoing challenges in the field is pulling meaningful information about animal movement from video footage. We either process videos manually, requiring many hours of tedious work, or focus on very simplistic and limited analysis that can be automated. The algorithms presented in this paper have potential to automate the labor-intensive part of our work more than has been possible previously, which could allow us to study a greater variety of animal locomotor behaviors.”
Once they have a database of motion and behaviors, the neuroscientists on the team can draw connections to the neural processes behind them. This will allow researchers “to not only gain a better understanding of how the brain produces behaviors,” said Shaevitz, “but also to explore future diagnostics and therapies that rely on a computer interpreting someone’s actions.”
A similar tool(Link is external) was shared over the summer by a team of Harvard researchers, who used existing neural network architecture, whereas the Princeton team created their own. “Our method and theirs have different advantages,” said Murthy. “This is an incredibly exciting field right now with a lot of activity in developing AI tools for studies of behavior and neural activity.”
“We use a different approach, where smaller, leaner networks can achieve high accuracy by specializing on new datasets quickly,” said Pereira. “More importantly, we show that there are now easy-to-use options for animal pose tracking via AI, and we hope that this encourages the field to begin to adopt more quantitative and precise approaches to measurement of behavior.”
“In the last five years, neuroscience has made enormous strides in the technology observing and manipulating brain activity,” said co-author Samuel Wang(Link is external), a professor of molecular biology and PNI. “Now, automatic classification of behavior adds a critical complement to that technology. Princeton is becoming a central hub in the budding field of computational neuroethology.”
“Fast animal pose estimation using deep neural networks(Link is external)” by Talmo Pereira, Diego Aldarondo, Lindsay Willmore(Link is external), Mikhail Kislin, Samuel Wang, Mala Murthy and Joshua Shaevitz was released online on Dec. 20 and published in the January 2019 issue of Nature Methods. The research was supported by the National Institutes of Health R01-NS104899-01 BRAIN Initiative Award, R01-MH115750 and R01-NS045193; the National Science Foundation through a BRAIN Initiative EAGER Award and GRFP DGE-1148900; the Nancy Lurie Marks Family Foundation; and a Howard Hughes Medical Institute Faculty Scholar Award.

The researchers on the project include (from left): Michail Kislin, a postdoctoral research associate; Lindsay Willmore, a graduate student; faculty members Joshua Shaevitz and Sam Wang; Talmo Pereira, a graduate student; and faculty member Mala Murthy.