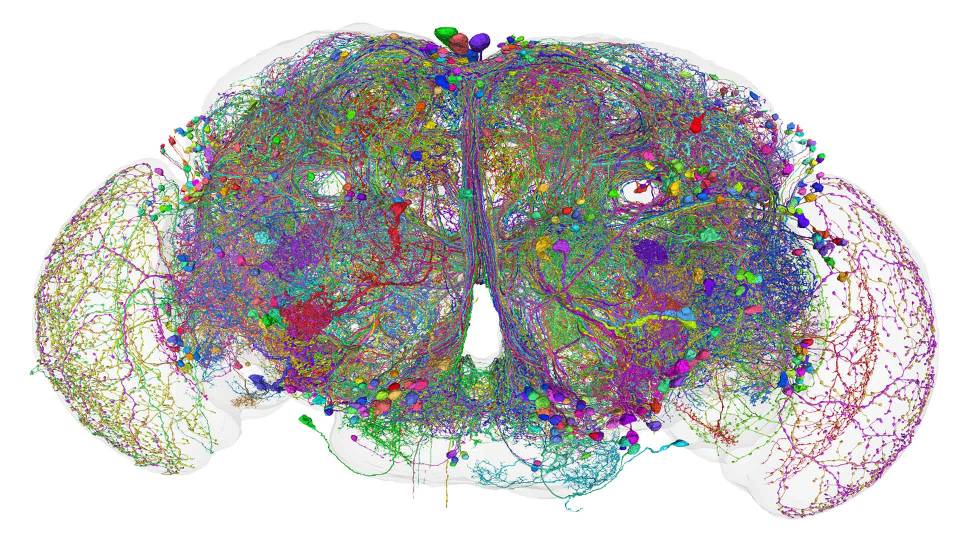
A Princeton-led team of researchers and gamers has mapped every neuron and synapse in the brain of an adult fruit fly.
For many heartbreaking diseases of the brain — dementia, Parkinson’s, Alzheimer’s and others — doctors can only treat the symptoms. Medical science does not have a cure.
Why? Because it’s difficult to cure what we don’t understand, and the human brain, with its billions of neurons connected by a hundred trillion synapses, is almost hopelessly complex.
“FlyWire,” a Princeton-led team of scientists and citizen scientists, has now made a massive step toward understanding the human brain by building a neuron-by-neuron and synapse-by-synapse roadmap — scientifically speaking, a “connectome” — through the brain of an adult fruit fly (Drosophila melanogaster). The FlyWire Consortium comprises members from more than 146 labs at 122 institutions, with major contributions from teams at the University of Cambridge and the University of Vermont.
“Any brain that we truly understand tells us something about all brains,” said Sebastian Seung, Princeton’s Evnin Professor in Neuroscience and a professor of computer science. “With the fly wiring diagram, we have the potential for an unprecedented, detailed and deep understanding.”
Previous researchers have mapped the brain of a C. elegans worm, with its 302 neurons, and the brain of a larval fruit fly, which had 3,000 neurons, but the adult fruit fly is several orders of magnitude more complex, with almost 140,000 neurons and tens of millions of synapses connecting them.
“This is a major achievement,” said Mala Murthy, director of the Princeton Neuroscience Institute and, with Seung, a leader of the research team. “There is no other full brain connectome for an adult animal of this complexity.”

FlyWire co-leaders Mala Murthy, the Karol and Marnie Marcin '96 Professor of Neuroscience and the director of the Princeton Neuroscience Institute, and Sebastian Seung, professor of computer science and the Evnin Professor in Neuroscience
The results of their work are featured in a special issue of the journal Nature about transformational research into the fruit fly brain. Seung and Murthy are co-senior authors on the flagship paper of the issue, which includes a suite of nine related papers with overlapping sets of authors, led by researchers from Princeton University, the University of Vermont, the University of Cambridge, the University of California-Berkeley, the University of Würzburg, Columbia University, UC-Santa Barbara, Freie Universität-Berlin, the University of Michigan, Johannes Gutenberg-Universität Mainz, and the Max Planck Florida Institute for Neuroscience.
A pathway toward tailored treatments
“How the brain functions depends critically on which neurons connect to which other neurons and the strength of their connections,” said Murthy, Princeton’s Karol and Marnie Marcin ‘96 Professor of Neuroscience. “To have a full wiring diagram of the fly brain — as a Drosophila neurobiologist, this is something I’ve dreamed of since I started my lab in 2010.”
Mala has pursued that vision — a full connectome of the fruit fly brain — since beginning her collaboration with Seung in 2018. “As neuroscientists, we have a habit of simplifying, of saying, ‘Hey, can I find that one neuron or cell type and figure out its function, how it contributes to animal decisions and behaviors? If I can simplify the problem down to that one neuron, then I can figure out what it does,’” said Murthy. “But when you take a step back, you realize ‘that one neuron’ is in a complex network of almost 140,000 neurons collectively coordinating this behavior.”
Neuroscientists like to point out that the human brain is the body’s most complex organ, and possibly the most complex neural network anywhere. “In many respects, it is more powerful than any human-made computer, yet for the most part we still do not understand its underlying logic,” said John Ngai, director of the U.S. National Institutes of Health’s BRAIN Initiative, which provided partial funding for the connectome project. “Without a detailed understanding of how neurons connect with one another, we won’t have a basic understanding of what goes right in a healthy brain or what goes wrong in disease.
“The collaborations across diverse areas of expertise in this type of team science consortium have brought the Drosophila brain map to light at an unprecedented pace, paving the way for detailed maps of the human brain and the tailored treatments that could follow,” Ngai said.
How gamers and AI helped make it happen
Sven Dorkenwald, the lead author on the flagship Nature paper, spearheaded the FlyWire consortium that mapped the fly brain.
“What we built is, in many ways, an atlas,” said Dorkenwald, a 2023 Ph.D. graduate of Princeton, now at the University of Washington and the Allen Institute. “Just like you wouldn’t want to drive to a new place without Google Maps, you don’t want to explore the brain without a map. What we have done is build an atlas of the brain, and added annotations for all the businesses, the buildings, the street names. With this, researchers are now equipped to thoughtfully navigate the brain, as we try to understand it.”
The map was built from 21 million images taken of the fruit fly brain by a team of scientists in Davi Bock’s lab, then at the Howard Hughes Medical Institute’s Janelia Research Campus. Using an AI model built by the Seung lab, the lumps and blobs in those images were turned into a labeled, three-dimensional map by the FlyWire Consortium — an unlikely collaboration among gamers, professional tracers, and neuroscientists who are collectively listed as last author on the flagship paper.
FlyWire took inspiration from the earlier EyeWire project, a crowdsourced gamer project that mapped neurons in a mouse retina. When EyeWire was launched, about 10 years ago, artificial intelligence hadn’t advanced to a point where it could accurately trace out each neuron, so gamers painstakingly assembled millions of tiny puzzles to solve the 3D structure of each mouse neuron, revealing each point of connection between them.
In the intervening decade, AI models trained on their work improved their ability to trace out neurons and synapses. Now humans serve as proofreaders, checking the AI-generated products and assembling the countless pieces into one massive whole, as well as annotators, adding cell type labels to each neuron. A team led by Gregory Jefferis at the MRC Laboratory of Molecular Biology and the University of Cambridge, and Bock, now at the University of Vermont, led the effort to add hierarchical annotations to all neurons in the connectome; their work appears in a companion paper in the special issue of Nature and completes the description of the FlyWire resource.
“FlyWire expanded on EyeWire not only in the AI improvements but also in the type of contributions that members could make,” said Amy Sterling, executive director of EyeWire and the crowdsourcing manager of FlyWire. “Members of EyeWire only mapped cells. Members of the FlyWire Consortium were able to both map neurons and contribute labels, both of which they did by the tens of thousands. Labs that originally competed ended up collaborating. Community members built apps and plug-ins to enhance the usage of FlyWire. Great beauty is revealed both in a map of a brain and in the idea that hundreds of human brains worked together make it all possible.”
This collaborative work was made possible through advances in computational infrastructure running in the cloud that the Seung and Murthy labs developed in collaboration with the Allen Institute for Brain Science. Since 2019, the researchers and gamers of FlyWire have collectively contributed 33 person-years to proofreading and annotating the results of the AI model. Without AI, Seung said, the project would have taken almost 50 thousand person-years.
“This dataset is a remarkable story of the power of open team science,” said Forrest Collman, associate director at the Allen Institute for Brain Science. “A dataset was produced and released by the Bock lab, picked up by researchers at Princeton, combined with open-source software to distribute the data to people spread across the world for proofreading, and then collaboratively analyzed by the Drosophila neuroscience community.”
Mapping the forest and the trees
The collaboration of online gamers, tracers, neuroscientists and cutting-edge artificial intelligence resulted in a map of every one of the fruit fly’s 139,255 neurons and 50 million of their synaptic connections. The word connectome highlights that it is those connections — synapses — between neurons where the brain’s most vital work takes place.
Most neurons look a bit like a tree, with a trunk, branches, roots and twigs. Just as a tree affects its neighbors, its roots connecting to surrounding organisms and its branches battling for sunlight, neurons connect with each other via synapses. But a whole brain is even more connected than a forest, because neurons can reach each other across comparatively massive distances.

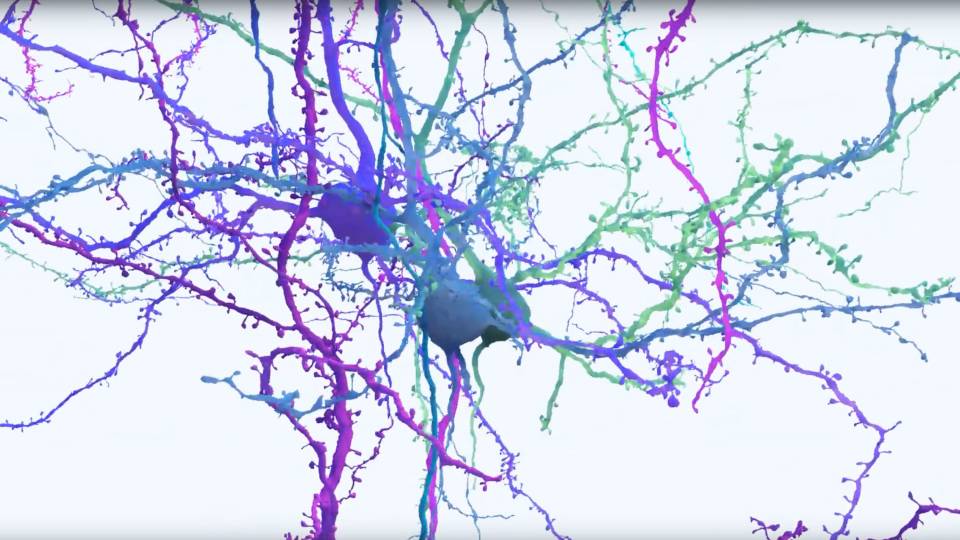
In a fruit fly brain, signals (seen here in pink) travel along neurons (blue) via many of the same neurotransmitters found in humans, including dopamine, glutamate and acetylcholine.
“It would be like a tree in New York interacting with a tree in Los Angeles,” said Dorkenwald. “Some of these neurons span the entire brain, from one eye to the other eye. There’s a diversity of sizes, from tiny neurons to others 100 times as big.”
And just like a map that traces out every tiny alley as well as every superhighway, the fly connectome shows connections within the fruit fly brain at every scale.
Once the connections were fully mapped, the team wanted to make it useful to the thousands of scientists conducting research in the field. To address this need, PNI research scientist Arie Matsliah, second author on the flagship paper, together with Sterling and a team of PNI developers, developed the FlyWire Codex. Matsliah calls it “Google for the connectome.” With the Codex, anyone with internet access can navigate every neuron and synaptic pathway in the brain map, without having to download massive amounts of data or knowing any advanced data analysis techniques. It has been already used by 10,000 people worldwide, with thousands of new searches processed daily.
The humble fruit fly

The common fruit fly is a simple creature with sophisticated behaviors, including flying, decision-making and serenading. As the subject of more than a century of research, fruit flies feature in countless scholarly articles detailing its physiology and development. Now that a Princeton-led team has traced every neuron in the brain of a fruit fly like this one, existing research can be linked to specific neural pathways and clusters.
“It might surprise people that flies have brains, but they do,” said Seung. “And their brains have neurons, and while their neurons don't look exactly the same as ours, they do look more or less like trees, like human neurons. It’s amazing. Our last common ancestor might have been half a billion years ago, and yet fruit flies have recognizable neurons and the same neurotransmitters we have: glutamate, acetylcholine, dopamine.”
Most of us don’t think about fruit flies unless they’re circling over our bananas. With a whole body that’s basically a speck, their brains are almost incomprehensibly tiny — about 750 microns across, 350 microns tall, and 250 microns deep. That’s significantly smaller than a poppy seed.
But this tiny insect has many behaviors that our larger, more complex bodies share, from complicated actions like communicating with romantic partners to simpler ones like moving rapidly, navigating, foraging for food, avoiding predators, responding to light and dark — indeed, fruit flies were responsible for the discovery of circadian rhythms, which spawned a whole field of brain and behavioral science.
Six different Nobel Prizes have honored 10 researchers studying D. melanogaster, including the 1995 Nobel Prize in Physiology or Medicine for Princeton’s Eric Wieschaus, who discovered the genes controlling embryonic development in fruit flies, and which were later found to be important in cancer research as well.
“Fruit flies are a wonderful model organism, because it’s quite small, but at the same time it has very complex behaviors,” Dorkenwald said. “As humans, we can relate to how male fruit flies sing to females, how they court them and follow them, and the competition between them.” Fruit fly behavior and physiology have been studied extensively for more than a century, so researchers have a wealth of pre-existing knowledge to tie to this new connectome of neurons and synapses.
‘A massive, interdisciplinary effort’
“This extraordinary accomplishment is the result of a massive, interdisciplinary team effort,” said Murthy. “We brought together Drosophila neuroscientists, with crowdsourced gamers and BRAIN Initiative funds and the ingenuity of our people here at Princeton.” The University’s endowment supported the effort via the Bezos Center for Neural Circuit Dynamics and the McDonnell Center for Systems Neuroscience.
At the University and across the nation and the world, the network of collaborators was vast. “At Princeton alone, we’ve had many postdocs and students working together with software engineers and full-time proofreaders,” Murthy said.
“Worldwide, there are more than 280 members in the FlyWire community building the connectome, most from the neuroscience and Drosophila science communities,” she continued. “And why are they motivated to do this for us? Because they can use the data for their own science.”
Instead of keeping their data confidential, the Princeton researchers opened their in-progress neural map to the scientific community from the beginning. “It took us about two years to correct all the errors,” said Murthy. “We released the data openly from Day One to the entire fly community and said, ‘Do whatever science you want, but help us proofread and annotate it as you go.’”
“Now that we have this brain map, we can close the loop on which neurons relate to which behaviors,” Dorkenwald said. “It’s wonderful, because new experiments will prompt new hypotheses, and we can relate things to the whole connectome, and we can iterate. I think the hard work is ahead. This is a beginning, not the end of the work.”

Why should we care about fruit flies?
- Fruit fly brains solve many of the same problems that we do, including figuring out how to navigate from point A to B, detecting scents in nature, and determining which way objects are moving in their environment.
- Fruit flies share 60% of human DNA, including genes for learning, Down syndrome and jet lag.
- 3 in 4 human genetic diseases have a parallel in fruit flies.
- Fruit fly research is responsible for six Nobel Prizes, including one won by Princeton’s Eric Wieschaus in 1995.
- Fruit flies age like we do, can get drunk, can be kept awake with coffee, and serenade their romantic interests.
- NASA sent fruit flies into space in 1947 — and they returned alive, paving the way for all astronauts.
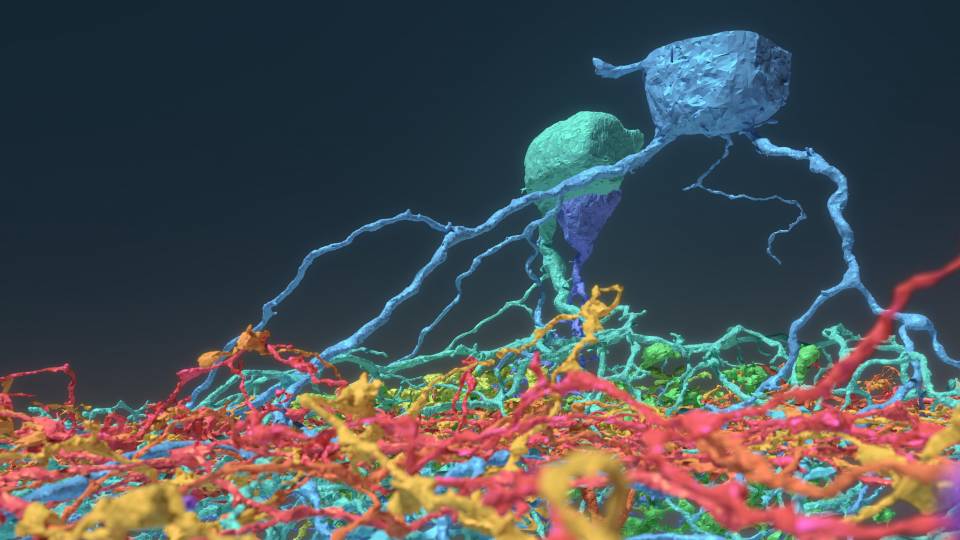
The accompanying image shows brain cells from the fruit fly’s auditory circuits, as mapped and annotated by FlyWire.
The Princeton research team includes Mala Murthy, the Karol and Marnie Marcin ’96 Professor of Neuroscience and the director of the Princeton Neuroscience Institute (PNI); Sebastian Seung, the Evnin Professor in Neuroscience and a professor of computer science; Sven Dorkenwald, a 2023 Ph.D. graduate who is now a Shanahan Fellow at the Allen Institute and the University of Washington; Arie Matsliah, a research scientist and data evangelist in PNI; Amy Sterling, a crowdsourcing specialist with PNI and the executive director of EyeWire; Szi-chieh Yu, a senior research specialist at PNI; Claire McKellar, a community manager for FlyWire; Albert Lin, a member of the Class of 2015 and now an associate research scholar at PNI and in Princeton’s Center for the Physics of Biological Function (CPBF), a National Science Foundation Physics Frontier Center; Chris Jordan and William Silversmith, full stack web developers with PNI; Akhilesh Halageri, a data infrastructure developer with PNI; Kai Kuehner, a research programmer with PNI; Oluwaseun (Seun) Ogedengbe, a former software developer with PNI now at LeanData; Ryan Morey, a research programmer at PNI; Jay Gager and Doug Bland, laboratory assistants in PNI; Runzhe (Tony) Yang, a 2023 Ph.D. in computer science and neuroscience now at SIG; David (Dudi) Deutsch, a former postdoctoral researcher at CPBF and now an assistant professor at the University of Haifa; Marissa Sorek, an EyeWire Game Master and image analyst with PNI; Ran Lu, a research scholar at PNI; Thomas Macrina, a 2022 Ph.D. in computer science; Kisuk Lee, a former postdoc at PNI now at Zetta AI; Alexander Bae, a 2022 Ph.D. in electrical and computer engineering; Shang Mu, a former associate research scholar in PNI; Barak Nehoran, a graduate student in computer science; Eric Mitchell, a member of the Class of 2018, now a graduate student at Stanford University; Sergiy Popovych, a 2022 Ph.D. in computer science now the CTO at Zetta AI; Jingpeng Wu, a former postdoctoral researcher now at ShanghaiTech University; Zhen Jia, a former postdoctoral researcher now at Amazon Web Services; Manuel Castro, a former research programmer; Nico Kemnitz, a former developer now at Zetta AI; and Dodam Ih, a former developer.
The full list of papers and their authors is available at Nature.
The flagship paper — Neuronal wiring diagram of an adult brain, by Sven Dorkenwald, Arie Matsliah, Amy R. Sterling, Philipp Schlegel, Szi-chieh Yu, Claire E. McKellar, Albert Lin, Marta Costa, Katharina Eichler, Yijie Yin, Will Silversmith, Casey Schneider-Mizell, Chris S. Jordan, Derrick Brittain, Akhilesh Halageri, Kai Kuehner, Oluwaseun Ogedengbe, Ryan Morey, Jay Gager, Krzysztof Kruk, Eric Perlman, Runzhe Yang, David Deutsch, Doug Bland, Marissa Sorek, Ran Lu, Thomas Macrina, Kisuk Lee, J. Alexander Bae, Shang Mu, Barak Nehoran, Eric Mitchell, Sergiy Popovych, Jingpeng Wu, Zhen Jia, Manuel Castro, Nico Kemnitz, Dodam Ih, Alexander Shakeel Bates, Nils Eckstein, Jan Funke, Forrest Collman, Davi D. Bock, Gregory S.X.E. Jefferis, H. Sebastian Seung, Mala Murthy and the FlyWire Consortium — appears in the current issue of Nature (DOI: 10.1038/s41586-024-07558-y ).
The research was supported by the National Institutes of Health (NIH) BRAIN Initiative (RF1 MH117815, RF1 MH129268, 1RF1MH120679-01 and U24 NS126935) and National Institute of Neurological Disorders And Stroke (NINDS) (RF1NS121911); the Princeton Neuroscience Institute’s Bezos Center for Neural Circuit Dynamics and McDonnell Center for Systems Neuroscience; Google; the Allen Institute for Brain Science; the National Science Foundation (NSF Neuronex2 2014862, Neuronex2 MRC MC_EX_MR/T046279/1, MRC MC-U105188491, PHY-1734030); Wellcome Trust Collaborative Award (203261/Z/16/Z and 220343/Z/20/Z); a Marie Skłodowska-Curie postdoctoral fellowship (H2020-WF-01-2018-867459); the Portuguese Research Council (Grant PTDC/MED-NEU/4001/2021); and the Intelligence Advanced Research Projects Activity (IARPA) via the Department of Interior (DOI) (D16PC0005).