Princeton University researchers are gaining new insights into the causes and characteristics of diseases by harnessing machine learning to analyze molecular patterns across hundreds of diseases simultaneously. Demonstrating a new tool now available to researchers worldwide, the team of computer scientists and biologists has already uncovered and experimentally confirmed previously unknown contributions of four genes to a rare form of cancer that primarily affects babies and young children.
The team, which includes collaborators at Michigan State University and the University of Oslo, introduced the system and demonstrated its abilities in a paper published in the Feb. 23 issue of the journal Cell Systems.
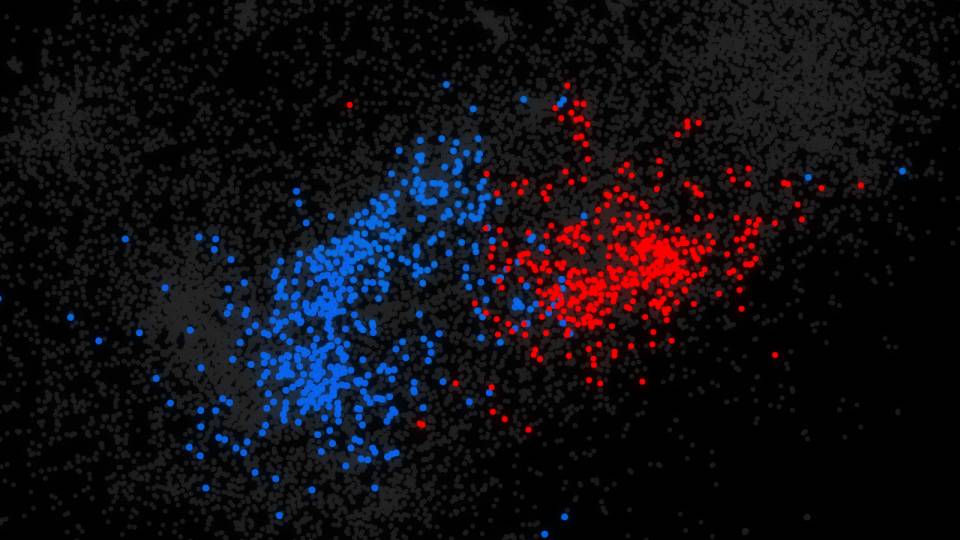
While previous approaches focused on genes associated with specific diseases or types of cancer, the new technique uses machine learning to find unique patterns of gene activity by looking at more than 300 different diseases simultaneously, including cancers, heart disease, metabolic disorders and many others. In doing so, it reveals distinctions between diseases and tissue types, including fine-tuned differences between related diseases that were not possible to discern with other techniques.

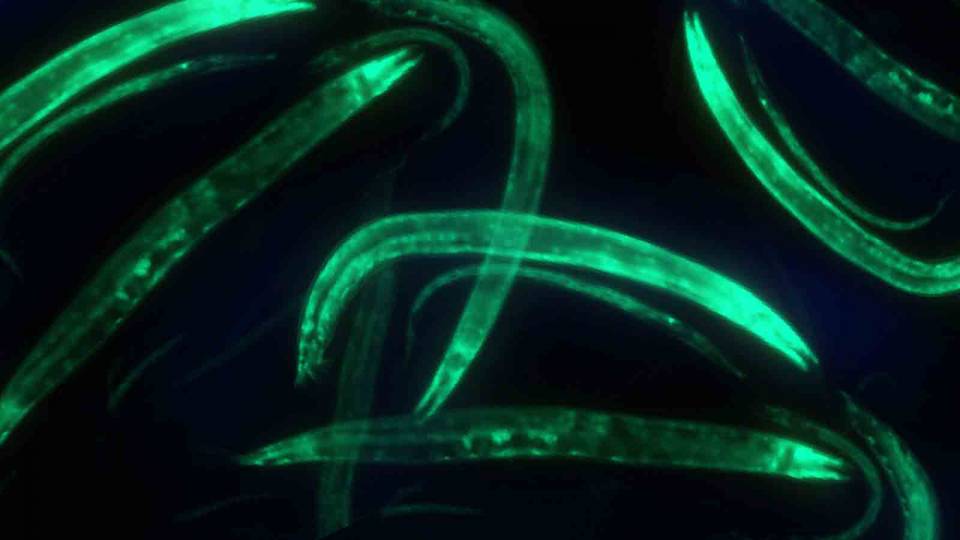
Princeton researchers created a data science tool that discerns fine-tuned distinctions between gene activity associated with specific diseases and tissues. In a demonstration of the tool’s power, the team discovered four genes associated with a rare pediatric cancer. The image on the left shows a normal cell while the one on the right highlights one of the discovered genes in neuroblastoma, which afflicts babies and young children.
The researchers believe that, with further development, the tool will be useful to clinicians in diagnosing disease, tailoring and tracking the effectiveness of therapies, and finding new treatment approaches.
The system, called Unveiling RNA Sample Annotation for Human Diseases, or URSA(HD), incorporates information about the activity of genes from publicly available records of about 8,000 biopsies taken from healthy and diseased tissues of thousands of patients. Going forward, researchers may submit new samples to the tool, via a web interface, and receive an analysis of possible associations with diseases and tissue types.
"The real innovation is comparing all samples to every other sample," said Chandra Theesfeld, one of the lead researchers along with Young-Suk Lee, who earned his Ph.D. at Princeton in 2016.
Theesfeld likened the idea to humans' ability to recognize nuanced differences between behaviors based on having seen a wide variety of examples. Watching soccer players, for example, might reveal the characteristics of a kicking action, but watching soccer players and ballet dancers at the same time reveals details and context for a similar action with a very different style and purpose.
"Studying them together provides a way to distinguish unique aspects," said Theesfeld, a research scientist in the lab of Olga Troyanskaya of Princeton and the Simons Foundation, who led the team. That viewpoint provides an unbiased way "to learn new things about disease that aren’t possible to find with the one-disease-at-a-time approach — and potentially identify new targets for therapies or even discover new aspects of disease that weren’t appreciated."
In making its comparisons, the algorithm gives more weight to differences in gene activity that uniquely define the distinct tissues and diseases. It de-emphasizes information about gene activity common to related diseases, much of which already is well studied. In the soccer-dancing analogy, it's like setting aside the large-scale action of lifting a leg in a kick and finding many details, such the angle of a foot, that taken together constitute a signature set of characteristics that reliably identifies one action or the other.
"Our method is driven by the disease information in the patient sample, so it’s not biased toward the popular disease genes that always get studied," Theesfeld said. "We can track patterns of changes in data without knowing exactly what each change means."
Theesfeld noted that 90 percent of studies of genes look at just 10 percent of human genes. URSA(HD) looks at the entire human genome and creates a genome-wide model or signature for each disease.
This approach could be particularly powerful for rare diseases, for which the researchers can now create a model with just a few samples. In the case of neuroblastoma, the pediatric cancer, the researchers found four genes that particularly contributed to the disease and for which there was no previous information in the scientific literature. To confirm the findings, Theesfeld performed laboratory tests on human cells, manipulating the gene activity and observing their effects on cancer-related processes in the cells.
Rather than looking at DNA itself, URSA(HD) looks at RNA, the product that cells create as they transcribe the information in DNA into working molecules that build and run cells and transmit signals from cell to cell. In this way, the system looks beyond mutations (scrambling in the genes themselves) and instead focuses on the downstream transcription products, which can be dysregulated in ways that cause problems even if the original gene is normal.
The research is part of longstanding work in Troyanskaya’s lab to integrate massive collections of dissimilar datasets to extract information necessary to make precise biological predictions and to direct laboratory experiments to accelerate discovery. A wide range of data science at Princeton brings together computing and biology to develop foundational tools and insights with potential to have a broad impact for health and humanity.
"Interdisciplinary approaches that merge sophisticated data science with deep knowledge of biology are key to deciphering biomedical puzzles necessary to realize the promise of precision medicine," said Troyanskaya.
In addition to Lee, Theesfeld and Troyanskaya, who is a professor of computer science and Lewis-Sigler Institute for Integrative Genomics and deputy director for genomics at the Flatiron Institute in New York, the researchers include: Rose Oughtred, Jennifer Rust, Christie S. Chang, Joseph Ryu and Kara Dolinski of Princeton; Arjun Krishnan of Michigan State University, and Vessela N. Kristensen of the University of Oslo. Support for the project was provided in part by the National Institutes of Health.