Red-hot rivers of molten copper and aluminum alloys streamed from one receptacle to another. As an undergraduate watching the demonstration in a materials science class, Clifford Brangwynne was reminded of cells migrating through the bloodstream. He realized at that moment that he could mold his interest in materials science and engineering into work that might ultimately have implications for human health.
Scientists like Brangwynne, now an assistant professor of chemical and biological engineering at Princeton, recognize the natural connection between engineering and the life sciences. Their research is setting the groundwork for future applications in health and medicine, including curing diseases such as Alzheimer's, growing replacement organs and preventing developmental abnormalities. Each of these pursuits hinges on the understanding that living matter obeys the same principles as nonliving matter.
Discovering the relevant principles — and using them to manipulate biological systems to meet our needs — is the goal of the growing field of bioengineering. "The thing that we do that's different from other scientists who are looking at states of matter and their properties is that we are doing it in the context of living cells," Brangwynne said. "What is the state of matter inside of a cell and how does that enable biological function?"
Brangwynne gestures toward a can of Gillette shaving foam next to a cylinder of silly putty on his desk and explains that the familiar grade school schema of three states of matter — solid, liquid, gas — is not entirely accurate. There are phases in between, and combinations with their own surprising properties.
"A mound of foam is essentially a solid," Brangwynne said. "You can push on it and it deforms, and when you take your finger away it springs back into shape. You've taken something that is 95 percent gas — it's mostly air — and 5 percent liquid, and you've combined those in such a way that you get a solid."
Correct biological function depends on transitions between these phases of matter. For example, your blood — typically a free-flowing liquid — clots to form a protective scab. However, these transitions can cause problems, for instance when an internal blood clot causes a stroke.
Researchers in the lab of Clifford Brangwynne, an assistant professor of chemical and biological engineering, use nucleoli from amphibian egg cells to study the role of gravity in determining the size of living cells. (Image courtesy of the Clifford Brangwynne lab)
Likewise, the liquid inside each cell — the cytoplasm — regularly goes through local phase transitions, some of which are disruptive. This issue is linked to neurodegenerative diseases, including Alzheimer's disease and amyotrophic lateral sclerosis (Lou Gehrig's disease). In these cases, proteins aggregate and spontaneously transition from a liquid phase into a sticky, solid-like state, prohibiting normal function in the brain or nervous system. These phase transitions are also thought to be involved in controlling cell size and growth, and thus diseases such as cancer.
Brangwynne's research focuses on gaining a better understanding of these living states of matter, and how they can be manipulated. Over the past few years, his group has published several studies exploring the molecules that control intracellular phase transitions and how the living matter within a cell affects gene regulation and cell size.
In one particularly exciting study, graduate student Marina Feric discovered that liquid-phase droplets of RNA and protein are biophysically linked to cell size through the force of gravity. Brangwynne, who receives support from the National Science Foundation (NSF) and the National Institutes of Health (NIH), is optimistic that these fundamental studies will lead to medical applications. "We're certainly hoping to use our findings to perturb these systems and keep cells in a healthier state," he said.
Swimming upstream
Like Brangwynne, Professor Howard Stone followed the flow of ideas from an area of engineering — fluid mechanics — to biology. "The living world involves flow almost by definition," said Stone, who is the Donald R. Dixon '69 and Elizabeth W. Dixon Professor of Mechanical and Aerospace Engineering. "You circulate blood, you breathe streams of air in and out, you sweat to regulate temperature. If you study fluid mechanics, and if you're somewhat open-minded, it's easy to stumble across biological problems."
One of the biological problems that caught Stone's attention is how bacteria move in a fluid. Moving through water is far more difficult for bacteria than it is for a human. For single-celled creatures a millionth of a meter long, the force of friction dominates their ability to swim in a given direction. Instead, bacteria are usually just carried along for the ride.
The dominance of friction led to a discovery in Stone's lab four years ago by visiting graduate student Yi Shen, who found that P. aeruginosa, a dangerous pathogen sometimes found in hospitals, can move against a current. The bacterium loses its flagellum — a long tail for swimming — when it adheres to a surface, which for many cells dictates the end of mobility. Yet P. aeruginosa can drag itself along a wall of, for example, a branched medical tube, by its small, tentacle-like pili, which are strong enough to resist the force of friction.

(Illustration by Matilda Luk, Office of Communications)
After observing this phenomenon, Stone and his collaborators — Professor of Molecular Biology Zemer Gitai; Albert Siryaporn, an associate research scholar in molecular biology; and Minyoung Kevin Kim, a graduate student in chemistry — began researching this behavior in systems mimicking the human body.
"Your blood vessels have branches. Your lungs are branched. We've made model branched systems and used a pump — like a heart — to drive fluid through it," Stone said. "Bacteria inoculated into this flow sometimes end up in places you wouldn't expect, and that, at this time, a simulation would never predict." This basic research, which is supported by NSF, may allow us to better anticipate the movements of pathogens — in our bodies or our environments — in order to prevent infection and contamination.
Fluid environment
Bacteria are single-celled organisms that can form colonies, but a more sophisticated arrangement occurs in our own bodies, where huge communities of cells organize into tissues and organs. Celeste Nelson, an associate professor of chemical and biological engineering, studies the fetal development of these organs, which depends on the fluid environment in which they form.
One of the organs that forms in a fluid environment is the lung. A fetus's lungs are filled with fluid during gestation. Nelson uses tissue cultures — parts of organs grown in laboratory dishes — and manipulates the speed and pressure of tiny streams of liquid that are directed onto the growing lung cells by small tubes.
Nelson has discovered that the higher the pressure of this fluid in the fetal lung, the more quickly the lungs develop, whereas lower pressure leads to slower development. Several congenital disorders can derail lung development, and Nelson's work — which is supported by NIH, NSF, the David and Lucile Packard Foundation, the Camille and Henry Dreyfus Foundation, the Burroughs Wellcome Fund, the Essig Enright Family Foundation, and Princeton's Project X, which provides seed funding for unconventional research — may improve our ability to diagnose such problems early.

Lung tissue extracted from a reptile embryo helps researchers in the lab of Celeste Nelson, an associate professor of chemical and biological engineering, study the effect of the fluid environment on lung development. (Image courtesy of the Celeste Nelson lab and exhibited in Princeton's 2011 "Art of Science" competition)
The lungs, kidneys, mammary glands and other organs develop through a branched structure, which is an efficient space-filling strategy for functions that require maximum surface area. This exponential branching pattern is a highly reproducible self-assembly process, and in Nelson's opinion, the forest of alveoli in the lungs is the most beautiful example. "The 23 generations of branches means several hundred million paths," said Nelson. Every one of those paths is needed for efficient diffusion of oxygen into the infant blood stream immediately after birth. "What’s amazing," she said, "is that all of the branches in my lungs look exactly like the branches in your lungs."
Nelson's lab also studies a behavior in cancer cells called reversion, which — if it could be induced — would turn many cancers into benign, treatable illnesses. She collaborates with Derek Radisky, a researcher at the Mayo Clinic in Jacksonville, Florida. For Nelson, who started studying breast cancer while a postdoctoral fellow, the body's organ systems have a mechanical elegance.
Timing is everything
Stanislav Shvartsman is as fascinated by the chemical aspects of development as Nelson is by the mechanical aspects. His research focuses on embryogenesis, the very early stages of fetal development.
"When you want to bake a cake, it’s not enough to say that you need eggs and milk and flour," said Shvartsman, a professor of chemical and biological engineering and the Lewis-Sigler Institute for Integrative Genomics. "Knowing the ingredients, and even knowing the sequence in which you add these ingredients — which is what we know from genetics — is not enough to bake a cake that tastes good."
When the recipe — the proper quantities of chemicals released by the cells of the embryo at the proper times — isn't followed exactly, there are consequences for the developing organism. For example, a large class of developmental abnormalities, known as RAS-opathies, is associated with asymmetry in the craniofacial complex, stunted height, congenital heart defects, developmental delays and other issues. Such defects are observed in one in every thousand births and are believed to be caused by mutations in genes of the Ras-MAPK pathway.
Biologists know which genes are mutated, and even where to find these genes on our DNA. What they don't know is why these particular mutations lead to a distinct set of clinical features. To find out, researchers turn to organisms that macroscopically look very different from us — such as bacteria and worms — but are very similar at the cellular level. Shvartsman's research group, which is supported by NIH and NSF, uses the fruit fly to study embryogenesis.
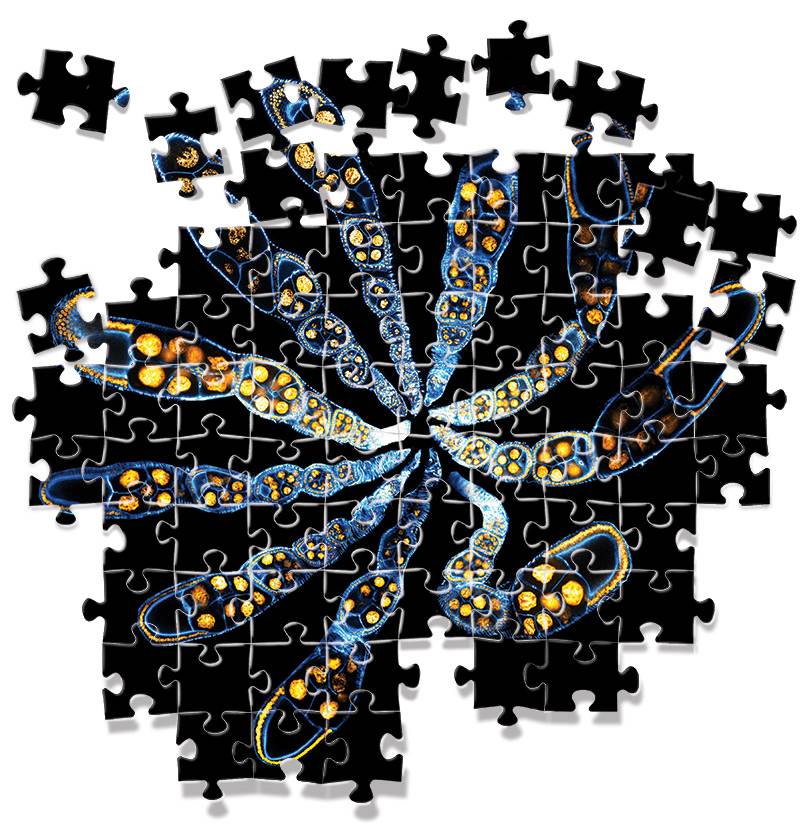
The image shows thin slices of a part of fruit fly embryos where stem cells turn into mature eggs. The image was created by graduate students Yogesh Goyal and Bomyi Lim and postdoctoral researcher Miriam Osterfield in the lab of Stanislav Shvartsman, a professor of chemical and biological engineering and the Lewis-Sigler Institute for Integrative Genomics. (Image courtesy of the Stanislav Shvartsman lab and exhibited in the 2014 "Art of Science" competition)
Initially, the handful of cells that make up an embryo are all identical. By the time of birth, that homogenous handful will have given rise to brain, nerve, heart, blood and every other kind of cell required for a living, breathing organism. In order to differentiate into the right kind of cell at the right time, and to arrange into the correct three-dimensional shape, the embryonic cells have to communicate. They speak to each other through a language of chemical signals.
The signals are actually protein molecules, Shvartsman said. A protein released by one cell attaches to a receptor protein embedded in the surface of a neighboring cell. That surface protein reacts by changing shape, and in turn changing the internal environment of its cell. In this way cells "hear" each other. At any given time multiple cells are releasing various proteins, and the combination of signals floating through the embryonic environment tells a cell what to become, or when to divide to make more of itself.
To crack the code, Shvartsman is looking at one signal at a time, beginning with a set of proteins that is well understood genetically thanks to the work of such Princeton biologists as Gertrud Schüpbach, the Henry Fairfield Osborn Professor of Biology. By controlling the amount of these proteins released in the fly embryo, Bomyi Lim, a former graduate student in Shvartsman's lab and now a postdoctoral research associate at the Lewis-Sigler Institute for Integrative Genomics, has discovered the minimum dosage necessary for proper structural development. This is the first step in a long process, but it is a milestone, and Shvartsman is excited about continuing the process. "It's very exciting to work in a field where there's no risk of ever saying, 'This is the end of the times-table. There is no more material to learn,'" he said.
Some assembly required
Brangwynne views embryogenesis as the epitome of self-assembly, the process by which small, disorganized components interact based on simple rules to form complex structures without human intervention. A classic example is the snowflake, a delicate crystalline jewel formed in midair as water molecules freeze. Engineers have been trying to take advantage of self-assembly for some time — often in Brangwynne's field of materials science — where time and money could be saved if certain synthetic materials would form on their own in a solution, rather than being painstakingly put together atom by atom.
"The embryo of an organism like C. elegans essentially starts out as just a bag of molecules," Brangwynne said. Once the egg is fertilized, it begins to organize, and the unstructured soup of molecules turns into a wriggling worm. "There's absolutely nothing that human engineering can do that comes anywhere close to what I just described takes place in embryos all the time," Brangwynne said.
While Princeton scientists are importing methods and paradigms from engineering disciplines to biology, they see a two-way street, recognizing that biology itself has methods to share.

"We would like to learn how nature, through hundreds of millions of years of evolution, has generated these systems that are just completely unbelievable in their level of sophistication," Brangwynne said. "It's as if we've been visited by an alien civilization that was millions of years more advanced than us. The first thing we would do would be to take a really close look at that spaceship. We'd try to figure out what it is made of, what are the principles that govern its flight and its control systems. That's what we are doing with biological systems."
This article was originally published in the University's annual research magazine "Discovery: Research at Princeton."