Life can be so intricate and novel that even a single cell can pack a few surprises, according to a study led by Princeton University researchers.
The pond-dwelling, single-celled organism Oxytricha trifallax has the remarkable ability to break its own DNA into nearly a quarter-million pieces and rapidly reassemble those pieces when it's time to mate, the researchers report in the journal Cell. The organism internally stores its genome as thousands of scrambled, encrypted gene pieces. Upon mating with another of its kind, the organism rummages through these jumbled genes and DNA segments to piece together more than 225,000 tiny strands of DNA. This all happens in about 60 hours.
The organism's ability to take apart and quickly reassemble its own genes is unusually elaborate for any form of life, explained senior author Laura Landweber, a Princeton professor of ecology and evolutionary biology. That such intricacy exists in a seemingly simple organism accentuates the "true diversity of life on our planet," she said.
"It's one of nature's early attempts to become more complex despite staying small in the sense of being unicellular," Landweber said. "There are other examples of genomic jigsaw puzzles, but this one is a leader in terms of complexity. People might think that pond-dwelling organisms would be simple, but this shows how complex life can be, that it can reassemble all the building blocks of chromosomes."
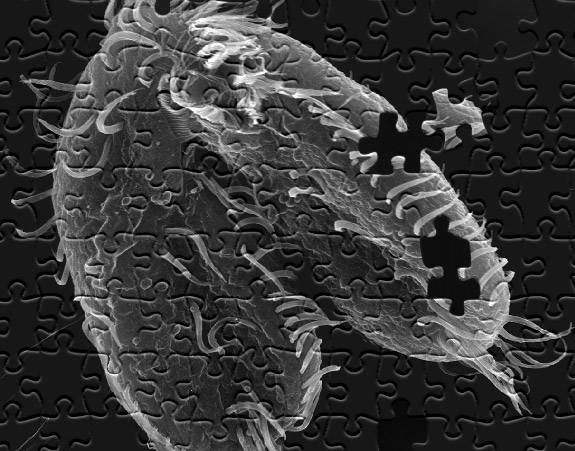
A study led by Princeton University researchers found that the pond-dwelling, single-celled organism Oxytricha trifallax (above) has the remarkable ability to break its own DNA into nearly a quarter-million pieces and rapidly reassemble those pieces when it's time to mate. This elaborate process could provide a template for understanding how chromosomes in more complex animals such as humans break apart and reassemble, as can happen during the onset of cancer. (Image by John Bracht, American University, and Robert Hammersmith, Ball State University)
From a practical standpoint, Oxytricha is a model organism that could provide a template for understanding how chromosomes in more complex animals such as humans break apart and reassemble, as can happen during the onset of cancer, Landweber said. While chromosome dynamics in cancer cells can be unpredictable and chaotic, Oxytricha presents an orderly step-by-step model of chromosome reconstruction, she said.
"It's basically bad when human chromosomes break apart and reassemble in a different order," Landweber said. "The process in Oxytricha recruits some of the same biological mechanisms that normally protect chromosomes from falling apart and uses them to do something creative and constructive instead."
Gertraud Burger, a professor of biochemistry at the University of Montreal, said that the "rampant and diligently orchestrated genome rearrangements that take place in this organism" demonstrate a unique layer of complexity for scientists to consider when it comes to studying an organism's genetics.
"This work illustrates in an impressive way that the genetic information of an organism can undergo substantial change before it is actually used for building the components of a living cell," said Burger, who is familiar with the work but had no role in it.
"Therefore, inferring an organism's make-up from the genome sequence alone can be a daunting task and maybe even impossible in certain instances," Burger said. "A few cases of minor rearrangements have been described in earlier work, but these are dilettantes compared to [this] system."
Burger added that the work is "extremely comprehensive as to the experimental techniques employed and analyses performed." The project is one of the first complex genomes to be sequenced using Pacific Biosciences (PacBio) technology that reads long, single molecules.
Oxytricha already stands apart from other microorganisms, Landweber said. It is a large cell, about 10 times the size of a typical human cell. The organism also contains two nuclei whereas most single-celled organisms contain just one. A cell's nucleus regulates internal activity and, typically, contains the cell's DNA as well as the genes that are passed along during reproduction.
An individual Oxytricha cell, however, keeps its active DNA in one working nucleus and uses the second to store an archive of the genetic material it will pass along to the next generation, Landweber said. The genome of this second nucleus — known as the germ-line nucleus — undergoes the dismantling and reconstruction to produce a new working nucleus in the offspring.
Oxytricha uses sex solely to exchange DNA rather than to reproduce, Landweber said — like plant cuttings, new Oxytricha populations spawn from a single organism. During sex, two organisms fuse together to share half of their genetic information. The object is for each cell to replace aging genes with new genes and DNA parts from its partner. Together, both cells construct new working nuclei with a fresh set of chromosomes. This rejuvenates them and diversifies their genetic material, which is good for the organism, Landweber said.
"It's kind of like science fiction — they stop aging by trading in their old parts," she said.
It's during this process that the scrambled genes in the germ-line nucleus are sorted through to locate the roughly 225,000 small DNA segments that each mate uses to reconstruct its rejuvenated chromosomes, the researchers found. Previous work in Landweber's lab — a 2012 publication in Cell and a 2008 paper in the journal Nature — showed that millions of noncoding RNA molecules from the previous generation direct this undertaking by marking and sorting the DNA pieces in the correct order.
Also impressive is the massive scale of Oxytricha's genome, Landweber said. A 2013 paper from her lab in PLoS Biology reported that the organism contains approximately 16,000 chromosomes in the active nucleus; humans have only 46. Most of Oxytricha’s chromosomes contain just a single gene, but even those genes can get hefty. A single Oxytricha gene can be built up from anywhere between one to 245 separate pieces of DNA, Landweber said.
The exceptional genetics of Oxytricha protect its DNA, so that mainly healthy material is passed along during reproduction, Landweber said. It's no wonder then that the organism can be found worldwide munching on algae.
"Their successful distribution across the globe has something to do with their ability to protect their DNA through a novel method of encryption, then rapidly reassemble and transmit robust genes across generations," Landweber said.
Landweber worked with, from her lab, first author Xiao Chen and Derek Clay, two graduate students in molecular biology, co-first author John Bracht and Aaron Goldman, postdoctoral fellows in ecology and evolutionary biology, and David Perlman, director of the molecular biology mass spectrometry facility. Other researchers on the paper are from the University of South Florida, the University of Bern in Switzerland, Indiana University, the Benaroya Research Institute at Virginia Mason in Seattle, and the Icahn School of Medicine at Mount Sinai.
The paper, "The architecture of a scrambled genome reveals massive levels of genomic rearrangement during development," was published Aug. 28 in Cell. The work was supported by the National Institutes of Health (grant nos. GM59708 and GM109459), the National Science Foundation (grant nos. 0900544 and 0923810), an NIH fellowship (1F32GM099462), and a NASA postdoctoral fellowship.